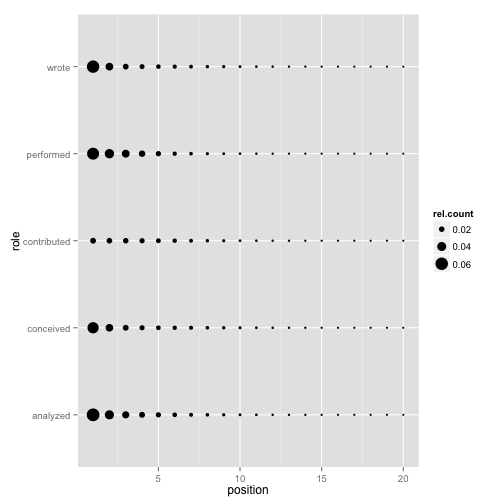
Position against contribution
This was the result of a Hack4ac team on 6th July 2013 working to mine and analyse article data from the PLOS Search API and PeerJ for detailed author contributions.
Several aspects were developed simultaneously:
- A PeerJ integration to pull author contribution information;
- A PLOS API bridge that queries the PLOS API, enriches it with author contribution information and converts it to CSV on the fly;
- A purely R-based implementation of mining PLOS for author contributions and a subsequent statistical analysis. This project is described in more detailed here and here.
PLOS API Bridge
The PLOS API bridge can be found under plos-api and is written in Ruby with Sinatra. To run it, ensure you set a PLOS_API_KEY environment variable like so:
$ export PLOS_API_KEY=decafbad
Then you can run the web application with Thin (as it streams responses):
$ bundle install
$ bundle exec thin start
Then to return CSV from PLOS, simply hit the root of the application:
GET /
DOI,Publication Date,Author Position,Author Name,Initials,Conceived and designed the experiments,Performed the experiments,Analyzed the data,Contributed reagents/materials/analysis tools,Wrote the paper
10.1371/journal.pone.0020946,2011-06-14T00:00:00Z,1,Olga Pantos,OP,1,1,1,1,1
10.1371/journal.pone.0020946,2011-06-14T00:00:00Z,2,Ove Hoegh-Guldberg,OH-G,0,0,0,1,0
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,1,Xiaowen Liu,XWL,1,1,1,0,1
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,2,Hong Cai,HC,1,0,0,1,0
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,5,Yingqiang Shi,YQS,1,0,0,1,0
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,6,Yanong Wang,YNW,1,0,0,1,0
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,4,Ziwen Long,ZWL,0,0,1,0,0
10.1371/journal.pone.0029670,2011-12-28T00:00:00Z,3,Hua Huang,HH,0,0,1,0,0
You can tweak the number of results returned by passing a limit parameter like so:
GET /?limit=1000
Datasets
Prepared datasets can be found under the data directory and are all in CSV format with descriptive headers.
Team Members

(Clockwise from bottom left.)